I'm interested in problems that are multidisciplinary by nature and that require creative and analytical thinking. My PhD work involved the mathematical analysis of 3D volumes of cardiac images as a means for visualizing and better understanding biological structures and processes in the heart. I have worked with a variety of methods originating from computer vision, computer graphics, and machine learning. In parallel of my research, I developed a 3D visualization and reconstruction engine for cardiac volumes in Java and OpenGL.
Research Interests
Documents
- PhD thesis - The Geometry of Cardiac Myofibers (PDF, 72MB)
- MSc thesis - Generalized Helicoids for Hair Modeling (PDF, 25MB)
- Comprehensive Exam - Neurovasculature Segmentation (PDF, 2MB)
- COMP 765 Report - A Pseudo-Physical Framework for Mapping Outdoor Terrains (PDF, 10MB)
- ECSE 626 Report - Image Registration by Maximization of Mutual Information (PDF, 1MB)
Publications
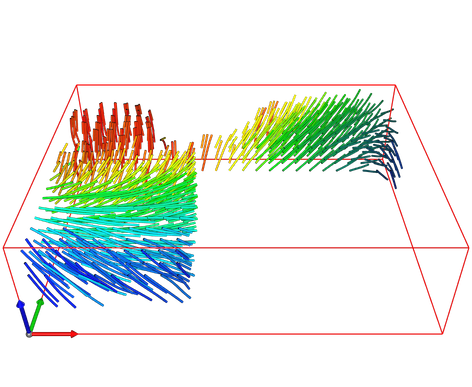 |
Moving Frames for Heart Fiber Reconstruction International Conference on Information Processing in Medical Imaging (IPMI). Sabahl Mor Ostaig, Scotland. 2015
|
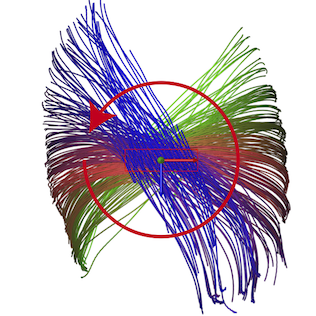 |
Maurer-Cartan Forms for Fields on Surfaces: Applications to Heart Fiber Geometry IEEE Transactions on Pattern Analysis and Machine Intelligence (PAMI). 2015
|
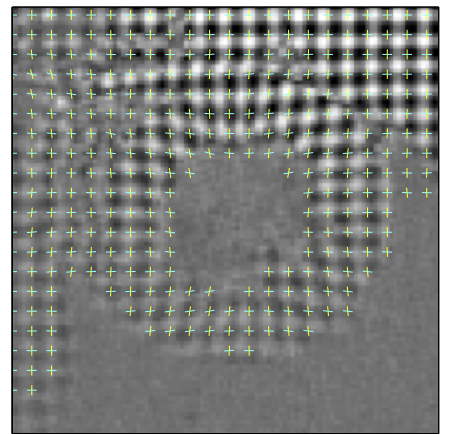 |
Connection Forms for Beating the Heart Statistical Atlases and Computational Models of the Heart (STACOM). Boston, USA, 2014
|
 |
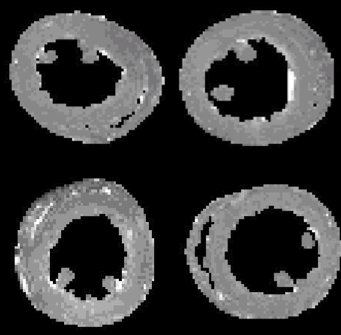
Cardiac Fiber Inpainting Using Maurer-Cartan Forms International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI). Nagoya, Japan, 2013
|
 |
Atlases of Cardiac Fiber Differential Geometry International Conference on Functional Imaging and Modeling of the Heart (FIMH). London, UK, 2013
|
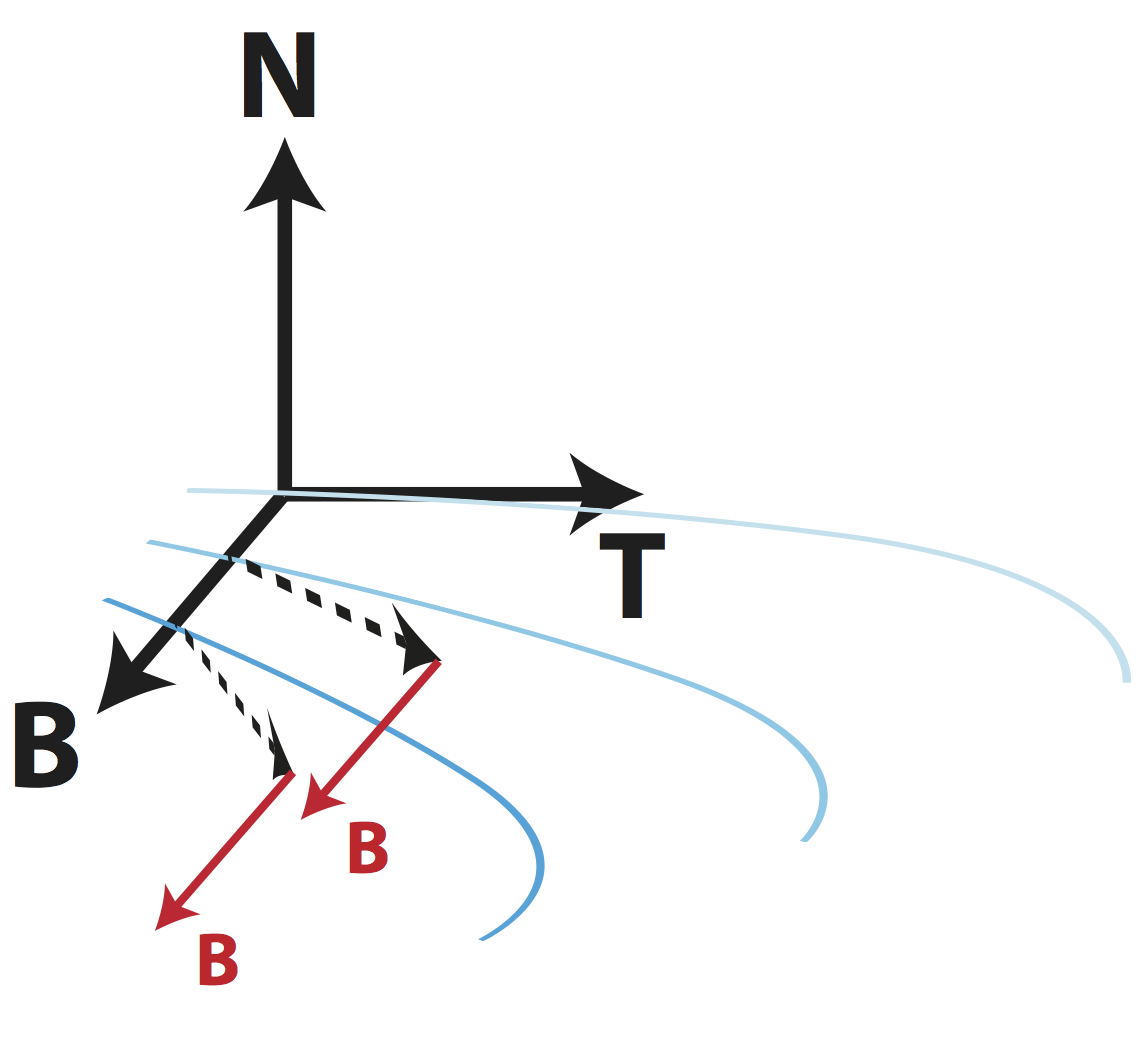 |
Moving Frames for Heart Fiber Geometry
International Conference on Information Processing in Medical Imaging (IPMI). Asilomar, USA, 2013.
|
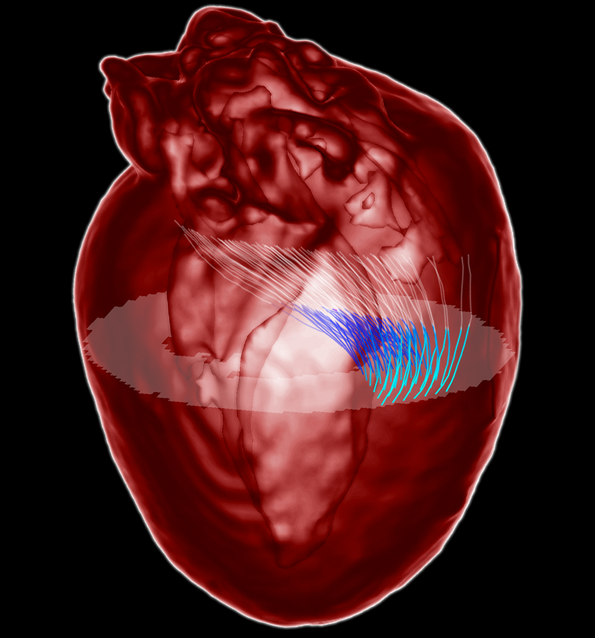 |
Heart wall myofibers are arranged in minimal surfaces to optimize organ function
Proceedings of the National Academy of Science (PNAS). 2012
|
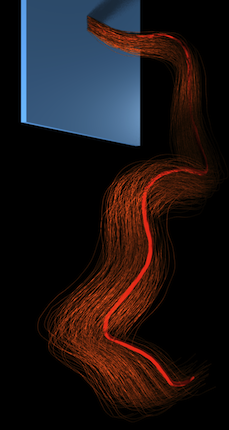 |
Generalized Helicoids for Modeling Hair Geometry
Proceedings of Eurographics (Computer Graphics Forum). 2011
|
Selected Past Research and Projects
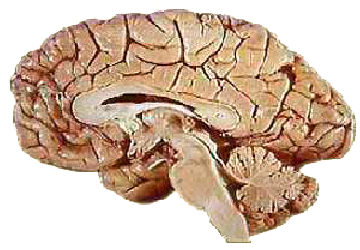 |
Phd Report - Neurovascular Segmentation
The identification of cerebral vessel lesions and malformations in medical images strongly correlates with risk factors for arterial embolisms (vascular blockage), ischemic strokes, and aneurysms, all of which are relatively common con- ditions that can be debilitating and fatal. Segmentation methods can support the clinician in extracting neurovasculature from medical images. These vascular representations are helpful in the development of safer treatments for cerebral conditions, for a more efficient preoperative planning, operating room and postoperative monitoring, and for intra- and inter-patient comparative studies. Common features of interest include vessel branch points, quantification of the spatial relationship between vessels, vessel segment extraction, vessel diameter measurements, and vessel complexity assessment. When a sequence of images is available, it can also be useful to quantify changes in these quantities over time or across patients, thereby opening up a lot of research potential. The main focus of this literature review is to present a selection of key articles covering different approaches employed in the vessel segmentation literature.
|
 |
M.Sc Thesis in Computer Science (Computer Graphics)
Hair-like patterns, comprised of dense collections of curve-like elements, are ubiquitous in our world. Consider examples arising in nature such as hair, fur, grass, feathers, and white matter fiber tracts in the brain, or those arising in man-made structures, such as textiles, optical fibers or threads. This thesis investigates an approach for generating, fitting, and interpolating such patterns. An emphasis is placed on hair modeling in computer graphics as the driving application for the methods proposed. Many intricacies of hair modeling are shared with those of fur, feathers, and grass modeling, and so the proposed methods translate to these and related cases as well.
|
 |
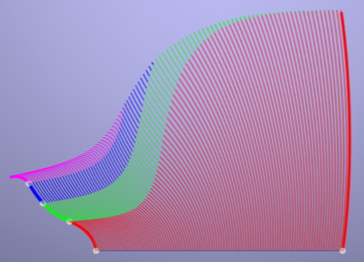
A Pseudo-Physical Framework for Mapping Outdoor Terrains
This course was an introduction to the broad area of mobile robotics. Topics included environment mapping, localization and path planning.
The course project consisted of a summary and/or suggested extensions to a selected research problem.
I designed and implemented a pseudo-physical mapping framework from the ground up in Java, which included implicit fitting and visualization, energy minimization, curvature and stationary point estimation and numerical derivation and integration.
Java OpenGL was used for rendering.
|
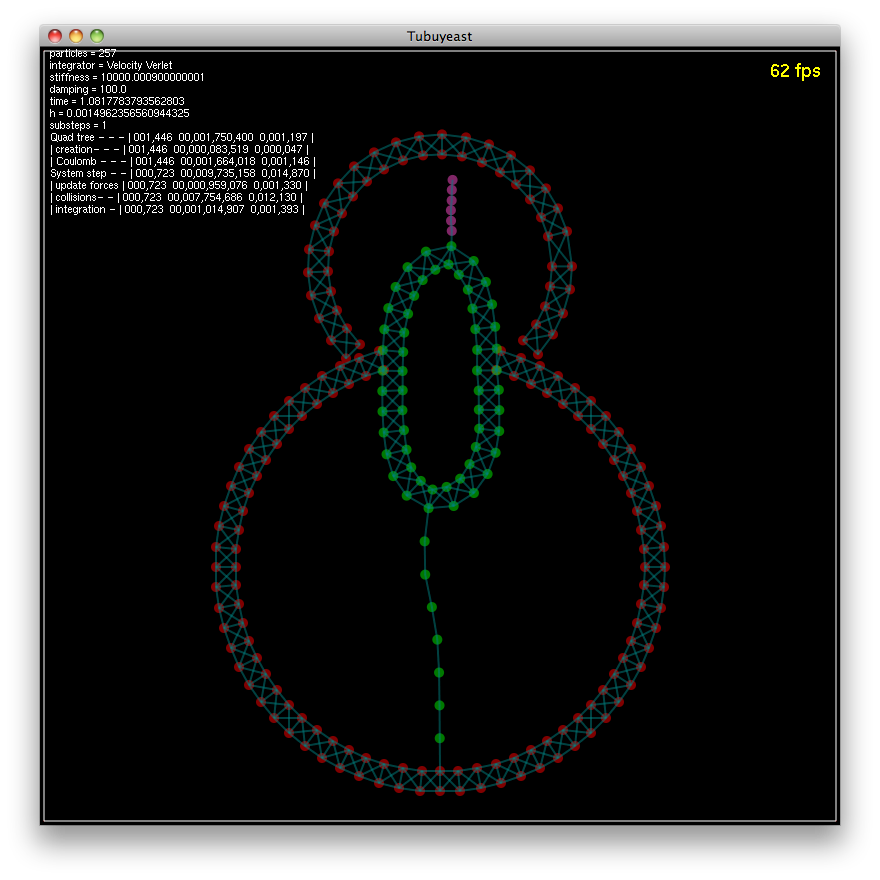 |
A Physical Simulation of Tubulin Dynamics in Yeast Cell Division
This course was an introduction to mathematical and computational methods in structural biology, and in algorithmic foundations of the approaches used in this field. The course project consisted of building a physical simulation of the dynamics of microtubules involved in yeast cell division. The framework was developed in Java/OpenGL.
|
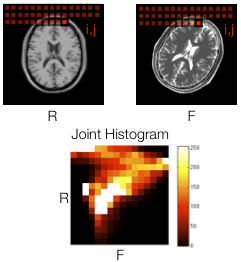 |
Multi-Modality Image Registration by Maximization of Mutual Information
The course project involved finding an interesting algorithm or
technique in the medical imaging literature, implementing it, and testing
it on a dataset. I picked a 1997 paper by Maes et al. describing one of the first
approaches using mutual information as a registration criterion in the
brain registration literature: Multi-Modality Image Registration by
Maximization of Mutual Information. The algorithm was implemented using
Matlab.
|
 |
Particle systems, cloth simulation, motion capture reuse
This course provided an introduction to computational
techniques for generating animation. Topics included physics-based
animation, implicit and explicit methods, collision detection and response, Lagrange
multiplier and energy constraints, motion capture, rigid
body motion, forward and inverse kinematics. My final project
consisted of implementing a hard constraint physical simulation: a particle
system cloth colliding with a sphere and sliding on a curtain-like
rail.
|
 |
Interactive Fine-tuning of Performance-driven Facial Animation
Facial animation is typically produced using a character rig, which
consists of a geometric model and a set of parametric deformations.
Motion capture can
greatly simplify the animation of the rig parameters, and has the
benefit of directly capturing all the subtle movements of a real face.
With motion capture, the problem is reduced to finding a mapping from the
captured performance to an existing rig. We present a method for interactively tuning motion
capture driven facial animation.
|